 |
Many files are associated with an RGS dataset, and it is easy to be overwhelmed.
The INDEX.HTM file, and links therein, are viewable with a web browser and
will help you navigate the dataset. The different types of files are discussed in
more detail in Chapter 1.
As ever, it is strongly recommended that you keep all reprocessed data in its own
directory! SAS places output files in whichever directory it is in when a task is
called. Throughout this primer, it is assumed that the Pipleline Processed data are
in the PPS directory, the ODF data (with upper case file names, and uncompressed) are
in the directory ODF, the reprocessing and analysis is taking place in the PROC directory,
and the CCF data are in the directory CCF.
If you have just received your data from the SOC, it has been processed with the
most recent version of SAS, and you should not need to repipeline it (though no
harm is done if you do); you need only to gunzip the files and prepare the data for
processing (see §5. However, it is very likely that you will want
to filter your data; in this case, you will need to reprocess it in order to determine the
appropriate filters. Therefore, we recommend that you rerun the pipeline regardless
of the age of your dataset.
But if you decide that reprocessing is unnecessary, you need only to gunzip the
files and rename event files for easier handling. For example, for the RGS1
event list,
where
As noted in Tables 3.2 and 3.3 you can view images
of your data. While the zipped FITS files may need to be unzipped before display
in ds9 (depending on the version of ds9), they can be displayed
when zipped using fv.
As usual, there are some HTML products to help you inspect the data.
These have file names of the form:
where
You will find a variety of RGS-specific files in XMM-Newton data sets. Generally
there are two of each because there are two RGS instruments. Table 3.3
lists typical file names, their purpose, the file format, and a list of tools that will
enable the user to inspect their data. Remember that the INDEX.HTM file
will help you navigate.
Various analysis procedures are demonstrated using the Mkn 421 dataset, ObsID 0153950701,
which definitely needs to be repipelined. The following procedures are applicable to all
XMM-Newton datasets, so it is not required that you use this particular dataset; any
observation should be sufficient.
We assume that the data was prepared and environment variables were set
according to §5, the GUI has been invoked (see
§5.3), and we are in our working directory, ``PROC''.
A window will appear with two panels. The upper one will
contain task names, what group they belong to (such as utility, epic, timing,
calibration, etc.) and a short description of each. The lower one will contain
information about environment variables, and as tasks are invoked, feedback
from the tasks.
From the upper window, select rgsproc. As with all SAS tasks in the GUI, double-clicking the task will bring up pop-up windows that will allow you to change the parameters. For rgsproc,
Note the last keyword, spectrumbinning. If you want to merge data from the same
orders in RGS1 and RGS2, keep it at the default value lambda. If you want to merge
data from the same instrument, with different orders, set it to beta. Merging spectra
is discussed in §11.7.
This takes several minutes, and outputs 12 files per RGS, plus 3 general use FITS files. At this point, renaming files to something easy to type is a good idea:
The pipeline task, rgsproc, is very flexible and can address potential pitfalls for RGS users. In §11.1, we used a simple set of parameters with the task; if this is sufficient for your data (and it should be for most), feel free to skip to later sections, where data filters are discussed. In the following subsections, we will look at the cases of a nearby bright optical source, a nearby bright X-ray source, and a user-defined source.
With certain pointing angles, zeroth-order optical light may be reflected off the
telescope optics and cast onto the RGS CCD detectors. If this falls on an
extraction region, the current energy calibration will require a wavelength-dependent
zero-offset. Stray light can be detected on RGS DIAGNOSTIC images taken before,
during and after the observation.
Please note that this will not work in every case. If a source is very
bright, the diagnostic data that this relies on may not have been downloaded
from the telescope in order to save bandwidth. Also, the RGS target itself
cannot be the source of optical photons, as the spectrum's zero-order falls
far from the RGS chip array.
This test, and the offset correction, are not
performed on the data before delivery.
To check for stray light and apply the appropriate offsets,
In the example above, it is assumed that the field around the source contains sky only. Provided a bright background source is well-separated from the target in the cross-dispersion direction, a mask can be created that excludes it from the background region. Here the source has been identified in the EPIC images and its coordinates have been taken from the EPIC source list which is included among the pipeline products. The bright neighboring object is found to be the third source listed in the sources file. The first source is the target:
If the true coordinates of an object are not included in the EPIC source list or the science proposal, the user can define the coordinates of a new source.
Since the event files are current, we can proceed with some simple analysis demonstrations, which will allow us to generate filters. Remember that all tasks should be called from the window where SAS was initiated, and that tasks place output files in whatever directory you are in when they are called.
The task xmmselect is used for many procedures in the GUI. Like all
tasks, it can easily be invoked by starting to type the name and pressing enter
when it is highlighted.
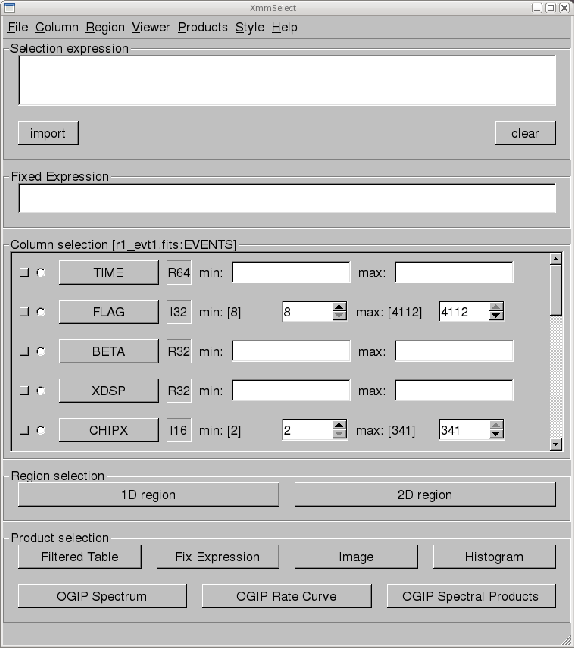
When xmmselect is invoked a dialog box will first appear requesting a
file name. You can either use the browser button or just type the file name
in the entry area, ``r1_evts1.fits:EVENTS'' in this case. To use the browser, select
the file folder icon; this will bring up a second window for the file selection.
Choose ``r1_evts1.fits'', then the ``EVENTS'' extension in the right-hand column,
and click ``OK''. The directory window will then disappear and you can click ``Run''
on the selection window.
When the file name has been submitted the xmmselect GUI (see Figure 11.1) will appear. (Later, when loading an event file that has been filtered, the filters that have been applied to that point will appear in the selection expression area.)
Two commonly-made plots are those showing PI vs. BETA_CORR (also known as ``banana plots'') and XDSP_CORR vs. BETA_CORR. To make a banana plot, invoke xmmselect in the SAS GUI and load the event file r1_evt1.fits, as discussed above. Then,
Different binnings and other selections can be invoked by accessing the ``Image'' tab at the top of the GUI. The default settings are reasonable, however, for a basic image. The resultant image is automatically displayed using ds9. Similarly, plots can be made comparing BETA_CORR to XDSP_CORR. These two example plots can be seen in Figure 11.2.
The background is assessed through examination of the light curve. We will extract
a region, CCD9, that is most susceptible to proton events and generally records the
least source events due to its location close to the optical axis. Also, to avoid
confusing solar flares for source variability, a region filter that removes the source
from the final event list should be used. The region filters are kept in the
source file product P*SRCLI_*.FIT. (For our example data, this would be
P0134520301R1S001SRCLI_0000.FIT).
More experienced users should be aware that with SAS 13, the *SRCLI* file's column information changed. rgsproc now outputs an M_LAMBDA column instead of BETA_CORR, and M_LAMBDA should be used to generate the light curve. (The *SRCLI* file that came with the PPS products still contains a BETA_CORR column if you prefer to use that instead.)
To create light curves, call xmmselect from the GUI. Then,
The resultant light curve is displayed automatically using Grace and is shown in Figure 11.3.
Examination of the lightcurve shows that there is a loud section at
the end of the observation, after 1.36975e8 seconds, where the count
rate is well above the quiet count rate of ![]() 0.05-0.2 count/second.
To remove it, we need to make an additional Good Time Interval (GTI) file and
apply it by rerunning rgsproc.
0.05-0.2 count/second.
To remove it, we need to make an additional Good Time Interval (GTI) file and
apply it by rerunning rgsproc.
There are two tasks that make a GTI file: gtibuild and tabgtigen.
Either will produce the needed file, so which one to use is a matter of the user's
preference. Both are demonstrated below.
The first method, using gtibuild, requires a text file as input.
In the first two columns, refer to the start and end times (in seconds) that
you are interested in, and in the third column, indicate with either a +
or - sign whether that region should be kept or removed. Each good (or bad) time
interval should get its own line. Comments can also be entered, if they are
preceeded by a "#". In the example case, we would write in our
ASCII file (named gti.txt):
and proceed to invoke the task gtibuild from the SAS GUI. Then,
Alternatively, we could filter on time using tabgtigen using the filtering
expression from the times noted previously. To do this, invoke tabgtigen
from the SAS GUI, and then
Finally, we could filter on rate using tabgtigen. The quiet
count rate for this observation is about 0.1-0.2 ct/s, so we will use that
in the filtering expression. Invoke tabgtigen from the SAS GUI,
and then
Now that we have GTI file, we can apply it to the event file by running rgsproc again. rgsproc is a complex task, running several steps, with five different entry and exit points. It is not necessary to rerun all the steps in the procudure, only the ones involving filtering. Call rgsproc from the SAS GUI, then
Since we made soft links to the event files, we don't need to rename them again.
Response matrices (RMFs) are now provided as part of the pipeline product package, but you might want create your own. The task rgsproc generates a response matrix automatically, but as noted in §11.1.4, the source coordinates are under the observer's control. The source coordinates have a profound influence on the accuracy of the wavelength scale as recorded in the RMF that is produced automatically by rgsproc, and each RGS instrument and each order will have its own RMF.
Making the RMF is easily done with the package rgsrmfgen. Please note that, unlike with EPIC data, it is not necessary to make ancillary response files (ARFS).
To make the RMFs, call rgsrmfgen from the GUI, then
RMFs for the RGS1 2nd order, and for the RGS2 1st and 2nd orders, are made in a similar way.
At this point, the spectra can be analyzed or combined with other spectra.
Spectra from the same order in RGS1 and RGS2 can be safely combined to create a spectrum
with higher signal-to-noise if they were reprocessed using rgsproc with
spectrumbinning=lambda, as we did in §11.1 (this also happens
to be the default). (Spectra of different orders, from one particular instrument, can also
be merged if the were reprocessed using rgsproc with
spectrumbinning=beta.) The task rgscombine also merges response
files and background spectra. When merging response files,
be sure that they have the same number of bins. For this example, we
assume that RMFs were made for order 1 in both RGS1 and RGS2 with rgsproc.
To merge RGS1 and RGS2 spectra, call rgscombine from the SAS GUI, and then
The spectra are ready for analysis, so we can now prepare the spectrum for fitting (§14).